Phenotypic and Molecular Characterization of the blaTEM Gene in Extended-Spectrum Beta-Lactamase-Producing Klebsiella pneumoniae
Abstract
Introduction
There has been a notable rise in antibiotic resistance among enterobacteria. This issue is primarily attributed to the emergence of extended-spectrum beta-lactamases (ESBLs), which present a significant concern for public health worldwide. This study investigates the prevalence of ESBL production, antibiotic resistance profiles, and molecular identification of the blaTEM gene in Klebsiella pneumoniae isolates.
Methods
The samples were randomly collected from several medical facilities in Erbil city. The VITEK 2 system was used for bacterial identification, antibiotic susceptibility, and ESBL production testing. The Double Disc Synergy Test (DDST) confirmed ESBL production. Polymerase chain reaction was conducted on all DNA samples, and the amplified DNA was analyzed using agarose gel electrophoresis to detect the blaTEM gene.
Results
A total of 43 samples were collected, of which the majority were urine (56%), followed by sputum (28%), blood (9%), and wound (7%). Klebsiella pneumoniae isolates exhibited the highest prevalence of resistance against ceftazidime (72%), ceftriaxone (70%), ciprofloxacin (63%), amoxicillin-clavulanic acid (60%), amikacin (58%), cefotaxime/tazobactam (56%), and gentamicin (53%). The DDST results indicated positive ESBL production in 15 isolates (35%), as evidenced by an increase or distortion in the inhibition zone toward the amoxicillin-clavulanate disc. Of the 43 isolates, 34 (79%) carried the blaTEM gene.
Conclusion
The study area shows a significant level of antibiotic resistance in ESBL-producing Klebsiella pneumoniae isolates, which, if not adequately addressed, could soon lead to severe health and therapeutic consequences.
Introduction
Antibiotic resistance is a dynamic and ongoing challenge. Over the past three decades, there has been a notable rise in antibiotic resistance among enterobacteria, particularly concerning third-generation cephalosporins. This issue is primarily attributed to the emergence of extended-spectrum beta-lactamases (ESBLs). ESBLs are a group of beta-lactamase enzymes produced by gram-negative bacteria that can hydrolyze and inactivate many antibiotics, including aztreonam, penicillin, and cephalosporins. These enzymes, such as TEM, SHV, and CTX-M, and their variants, allow enterobacteria to resist β-lactam antibiotics. The encroachment of these multi-drug-resistant pathogens into community settings raises significant alarm. The swift spread of the genes that encode these ESBLs, primarily via plasmids, has facilitated their rapid proliferation, leading to an alarming increase in the global prevalence of ESBL-producing bacteria. This situation presents a significant concern for public health worldwide [1,2]. Klebsiella pneumoniae is an opportunistic pathogen responsible for nosocomial and community-acquired infections [3]. This bacterium is usually found as an intestinal microflora. It can cause severe diseases like ventilator-associated pneumonia, catheter-related urinary tract infections, meningitis, bacteremia, septicemia, infection of surgical and non-surgical wounds, diarrhea, prosthetic valve endocarditis, peritonitis, and osteomyelitis [1]. The first strains of Klebsiella pneumoniae with ESBL (KP-ESBL) were identified in Europe in 1982, marking the emergence of resistance to ceftazidime and aztreonam due to plasmid-mediated beta-lactamase enzymes. These resistance traits rapidly disseminated to other gram-negative bacteria, including Escherichia coli. Since the discovery of these enzymes, their prevalence has continued to rise, with over 200 distinct ESBL enzymes identified today. The impact of ESBL-producing strains is particularly pronounced in intensive care units, which pose a high risk for epidemic outbreaks. K. pneumoniae and E. coli are the two most frequently encountered bacterial species associated with ESBL production. Implementing proactive monitoring of ESBL-producing pathogens in high-risk populations is crucial through suitable antimicrobial strategies. This necessity arises from the tendency of these pathogens to display multidrug resistance, which complicates treatment options and increases the potential for severe infections [4]. This study investigates the prevalence of ESBL production, antibiotic resistance profiles, and molecular identification of the blaTEM gene in Klebsiella pneumoniae isolates in Erbil City.
Methods
Study design and sample collection
This retrospective cross-sectional study was conducted between September 2021 and January 2022. The ethics board of the College of Health Sciences at Hewler Medical University approved the study. Patient consent was verbally obtained from the patients to collect their samples and be published in this study. The samples were randomly collected from several medical facilities in Erbil city, such as Nanakali Hospital, Hawler Teaching Hospital, and private laboratories. The collected samples included urine, sputum, or wound infection.
Isolation, identification, and susceptibility test of Klebsiella pneumoniae
The samples were cultured on MacConkey agar and incubated at 37°C for 24 hours. Klebsiella pneumoniae colonies were isolated based on their large, pink-to-red mucoid appearance. The VITEK 2 system (bioMérieux, France) was used for bacterial identification and antibiotic susceptibility testing. A sterile swab or stick was used to transfer an adequate number of colonies from the cultured sample into 3.0 ml of sterile saline (0.45% to 0.50% NaCl, pH 4.5 to 7) in a 12 × 75 mm polystyrene test tube. The suspension’s turbidity was then adjusted to match the 0.5 McFarland standard for antimicrobial susceptibility testing and measured using a DensiChek™ turbidimeter.
Phenotypic Detection and Confirmation of ESBLs
ESBL production was detected among the isolates using the VITEK 2 system. The testing panel included six wells containing 10 μg/ml ceftriaxone, 0.5 μg/ml cefotaxime, or 0.5 μg/ml ceftazidime, alone or in combination with clavulanic acid (10 μg/ml, 4 μg/ml, and 4 μg/ml, respectively). Bacterial growth in each well was quantitatively assessed using an optical scanner. A proportional reduction in growth in the wells containing cephalosporin combined with clavulanic acid, compared to those containing cephalosporin alone, was considered indicative of ESBL production. The Double Disc Synergy Test (DDST) confirmed ESBL production. Each Klebsiella pneumoniae isolate was inoculated on Müller-Hinton agar plates for susceptibility testing. Discs containing 30 μg of cefotaxime and ceftazidime were placed on either side of a disc with co-amoxiclav (20/10 μg), positioned 20 mm apart (center to center). ESBL production was indicated when the inhibition zone of either cephalosporin was expanded by clavulanic acid, often resulting in a distinctive "champagne-cork" or "keyhole" shape zone [1].
Molecular detection of blaTEM gene
DNA extraction of Klebsiella pneumoniae was carried out using the simple boiling method. Isolated colonies from overnight cultures were placed in a test tube containing 1 ml of distilled water and heated in a water bath for 10 minutes. The sample was centrifuged at 1,000 rpm for 5 minutes to complete the extraction process. Polymerase chain reaction (PCR) was conducted on all DNA samples of Klebsiella pneumoniae. The concentration of extracted double-stranded DNA was combined with a PCR master mix (Promega, USA), which included Taq DNA polymerase, deoxynucleotide triphosphates (dATP, dCTP, dGTP, dTTP), buffers, and salts. Specific forward (5'-GAG TAT CAA CAT TTC CGT GTC-3') and reverse (5'-TAA TCA GTG GGC ACC TTC TC-3') primers were added to amplify the blaTEM target gene. The PCR mixture was then incubated in a thermocycler, undergoing 32 cycles of alternating temperatures: a denaturation step at 94°C for one minute to separate the double-stranded DNA into single strands, an annealing step at 57°C for one minute to allow the primers to bind to the target DNA, and an extension step at 72°C for 10 minutes for the Taq DNA polymerase to synthesize new DNA strands. This process generated billions of copies of the target DNA sequence within a few hours. Subsequently, the amplified DNA was separated and analyzed using agarose gel electrophoresis.
Results
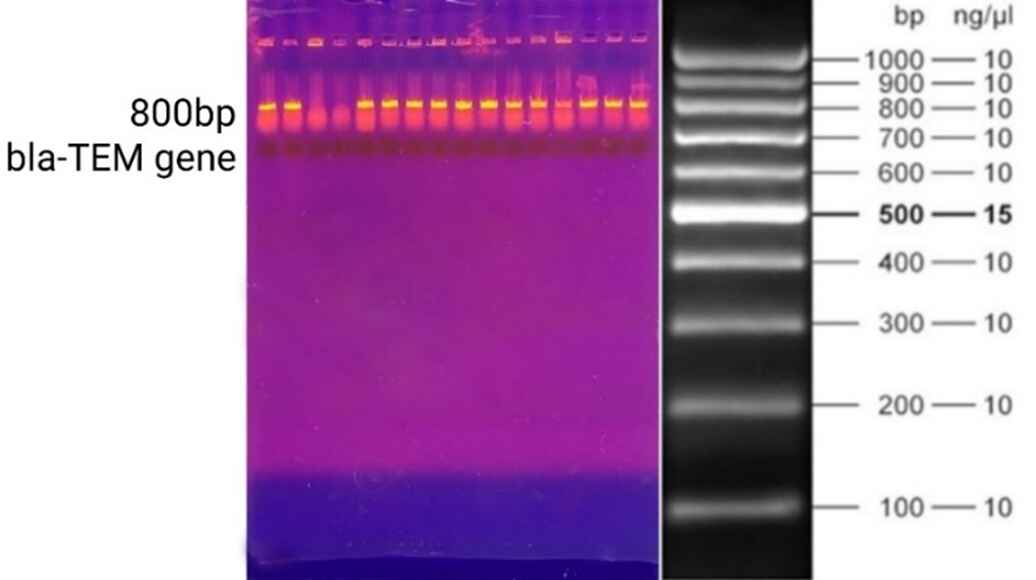
A total of 43 samples were collected; 22 (51%) were obtained from females and 21 (49%) from males. The majority of the samples were urine (56%), followed by sputum (28%), blood (9%), and wound (7%). Klebsiella pneumoniae isolates exhibited the highest prevalence of resistance against ceftazidime (72%), ceftriaxone (70%), ciprofloxacin (63%), amoxicillin-clavulanic acid (60%), amikacin (58%), cefotaxime/tazobactam (56%), and gentamicin (53%). In contrast, the lowest prevalence of resistance was observed with imipenem (26%) and meropenem (28%) (Table 1). The DDST results indicated positive ESBL production in 15 isolates (35%), as evidenced by an increase or distortion in the inhibition zone toward the amoxicillin-clavulanate disc. The remaining samples (65%) tested negative for ESBL production (Figure 1). Of the 43 isolates, 34 (79%) carried the blaTEM gene (Figure 2). Overall, 11 isolates (26%) tested positive in the DDST and PCR, while five isolates (12%) were negative. Additionally, four isolates (9%) were positive in DDST but negative in PCR, whereas 23 isolates (53%) were negative in DDST but positive in PCR (Table 2).

|
Antibiotic |
No. of resistant isolates |
Frequency (%) |
No. of sensitive isolates |
Frequency (%) |
|
Ceftazidime |
31 |
72% |
12 |
28% |
|
Ceftriaxone |
30 |
70% |
13 |
30% |
|
Ciprofloxacin |
27 |
63% |
16 |
37% |
|
Amoxicillin clavulanic acid |
26 |
60% |
17 |
40% |
|
Amikacin |
25 |
58% |
18 |
42% |
|
Cefotaxime |
24 |
56% |
19 |
44% |
|
Tazobactam |
24 |
56% |
19 |
44% |
|
Gentamicin |
23 |
53% |
20 |
47% |
|
Meropenem |
12 |
28% |
31 |
72% |
|
Imipenem |
11 |
26% |
32 |
74% |
| Isolated bacteria |
DDST results |
PCR results |
Number |
Frequency (%) |
|
(+) |
(+) |
11 |
26% |
|
|
(-) |
(-) |
5 |
12% |
|
|
(+) |
(-) |
4 |
9% |
|
|
(-) |
(+) |
23 |
53% |
Discussion
KP-ESBL has emerged as a significant concern due to its virulence factors and role as a leading cause of infectious diseases. It is categorized within the multidrug-resistant group of bacteria. Klebsiella pneumoniae exhibits antibiotic resistance through several mechanisms, including the enzymatic degradation or inactivation of antibiotic compounds, alterations in membrane permeability, and modifications of antibiotic target sites via bacterial protein mutations. It has been reported that K. pneumoniae acquires numerous ESBL enzymes, which serve as its primary defense mechanism against antibiotics [5]. The prompt and precise identification of ESBL-positive Enterobacteriaceae strains is critical for guiding appropriate patient management and developing and enforcing targeted infection control protocols [6]. Over the past decade, numerous studies have highlighted the prevalence of ESBL-mediated resistance in infectious bacteria, with a higher prevalence in Escherichia coli and Klebsiella pneumoniae [2,5,6-9]. In the study by Benbrahim et al., 15 out of 40 (37.5%) Klebsiella pneumoniae isolates were identified as ESBL-producing strains, a figure close to the 41.1% reported by Pirzaman et al. [10]. Another study reported a prevalence of 31.8% [2]. Higher rates were observed in Asian countries, reaching up to 75% [1]. In this study, the prevalence of isolates with positive ESBL enzymes was 35%, close to previously reported [1,2,10].
Infections caused by ESBL-producing bacteria can affect individuals of all ages, though their distribution may be influenced by patients' immunological status and the prevalence of antibiotic misuse. Benbrahim et al. found that KP-ESBLs were detected across all age groups, with the highest incidence (20%) observed in individuals aged 21-30 [1].
Gravey et al. reported a KP-ESBL prevalence of 4.1% in individuals aged 18–64 and 4.2% in those over 65 [11]. Due to the retrospective nature of the present study, the age preference among samples was unknown. In the present study, females comprised 51% of the sample, while males accounted for 49%, a distribution that closely aligns with the findings reported by Marra et al. [12]. However, a study by Ali and Ismael observed a higher proportion of female samples (72.72%) than male samples (22.73%) among 84 Klebsiella pneumoniae isolates [13]. Conversely, a study by Nirwati et al. found a higher frequency of male samples (64.07%) than female samples (35.93%) in 167 K. pneumoniae isolates [14]. In the study by Benbrahim et al., KP-ESBL strains were most frequently isolated from urine samples, which the authors suggested is likely due to urine being one of the most commonly collected specimens for clinical investigation [1]. Similarly, most of the samples (56%) in the present study were urine.
Antimicrobial resistance in pathogenic bacteria presents a global challenge, contributing to high mortality and morbidity rates. Infections caused by multidrug-resistant strains are often difficult or impossible to treat with conventional antimicrobials. The widespread, usually unnecessary use of antibiotics stems from the failure of many healthcare centers to promptly diagnose causative microorganisms and determine their antimicrobial susceptibility in patients with bacteremia and other serious infections [15]. In this study, Klebsiella pneumoniae strains exhibited the highest resistance rates against ceftazidime (72%), ceftriaxone (70%), ciprofloxacin (63%), and amoxicillin-clavulanic acid (60%). The resistance rate for ceftazidime in this study was slightly similar to that reported in another study conducted in Erbil, which documented a resistance rate of 62.5% [16]. Notably, our study found significantly higher resistance rates for ciprofloxacin (63% vs. 4.2%), gentamicin (53% vs. 4.2%), and amikacin (58% vs. 4.2%). Conversely, the resistance rate for amoxicillin-clavulanic acid was lower in our study than in the previous one (60% vs. 81.25%) [16]. Another study conducted in Iran reported even higher resistance rates for amoxicillin-clavulanic acid (100%), ceftazidime (84%), tazobactam (80%), ciprofloxacin (80%), and amikacin (60%) [17]. In contrast to our findings, sensitivity rates of 90% and 100% by K. pneumoniae isolates have been reported for amoxicillin-clavulanic acid and imipenem, respectively [1,18]. Muggeo et al. and Rasamiravaka et al. found a sensitivity rate of 83% to amikacin, while it was 42% in the present study. In the study by Bora et al., imipenem and meropenem were identified as one of the most effective antimicrobial agents against ESBL-producing isolates, comparable to the findings in this study [7].
The prevalence of the blaTEM gene in Klebsiella pneumoniae isolates varies across studies. Alibi et al. reported that blaTEM gene was present in 56.8% of their isolates [19]. In contrast, Paterson et al. found a prevalence of 87% [20]. Guessennd et al. reported that 63.4% of their isolates in Abidjan, Côte d’Ivoire, carried the blaTEM gene [21]. The prevalence rates of 31.57% and 55% for the blaTEM gene in Klebsiella pneumoniae have also been reported [2,5]. Based on genotypic detection, in this study, 79% of the isolates carried the blaTEM gene, which may indicate uncontrolled antibiotic misuse in our locality. In line with this finding, Bora et al. found that genes responsible for ESBL were positive in 79.45% of the Klebsiella pneumoniae isolates. However, in the phenotypic confirmatory test for ESBL production, 53.42% of the isolates tested positive, suggesting that the phenotypic test results may be subject to false positives or negatives. Several factors can contribute to this issue, including the production of multiple β-lactamase types by a single bacterial isolate, the co-production of ESBLs and constitutive AmpC β-lactamases, variations in substrate affinities, and the inoculum effect [7]. Accordingly, in the current study, phenotypic detection revealed that only 15 isolates (34.9%) were positive for ESBL production, while genotypic detection identified 34 isolates (79%) as positive for the blaTEM gene. Therefore, PCR using oligonucleotide primers specific to ESBL genes is considered the most straightforward and reliable method for detecting the presence of ESBLs [7].
This study has several limitations that may impact the generalizability of its findings, including small sample size, sampling restricted to a specific geographic area of the city, and the exclusive focus on the blaTEM gene. Future research with larger sample sizes and broader geographic coverage, extending beyond the city to the entire country, is needed to provide deeper insights into this issue.
Conclusion
The study area shows a significant level of antibiotic resistance in KP-ESBL strains, which, if not adequately addressed, could soon lead to severe health and therapeutic consequences. Molecular methods, which detect the genes responsible for resistance, may offer greater sensitivity in identifying antibiotic resistance.
Declarations
Conflicts of interest: The author(s) have no conflicts of interest to disclose.
Ethical approval: The ethics board of the College of Health Sciences at Hewler Medical University approved the study.
Patient consent (participation and publication): Written informed consent was obtained from patients for publication.
Source of Funding: Medical Research Center of Hawler Medical University.
Role of Funder: The funder remained independent, refraining from involvement in data collection, analysis, or result formulation, ensuring unbiased research free from external influence.
Acknowledgements: None to be declared.
Authors' contributions: MAG and KAO were significant contributors to the conception of the study and the literature search for related studies. LJM, LKHJ, and KAO were involved in the literature review, the study's design, and the critical revision of the manuscript, and they participated in data collection. HOA and KWM were involved in the literature review, study design, and manuscript writing. SYI and BMI Literature review, final approval of the manuscript, and processing of the tables. HOA and MAG confirm the authenticity of all the raw data. All authors approved the final version of the manuscript.
Use of AI: AI was not used in the drafting of the manuscript, the production of graphical elements, or the collection and analysis of data.
Data availability statement: Not applicable.
References
- Benbrahim C, Barka MS, Benmahdi L, Zatout A, Khadir A. Klebsiella pneumoniae producing extended spectrum β-lactamase in Regional Military University Hospital of Oran, Algeria: antibiotic resistance, biofilm formation, and detection of blaCTX-M and blaTEM genes. African Journal of Clinical and Experimental Microbiology. 2021 ;22(1):28-37. doi:10.4314/ajcem.v22i1.5
- Ugbo EN, Anyamene CO, Moses IB, Iroha IR, Babalola OO, Ukpai EG, et al. Prevalence of blaTEM, blaSHV, and blaCTX-M genes among extended spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae of clinical origin. Gene Reports. 2020; 21:100909. doi:10.1016/j.genrep.2020.100909
- Markovska R, Stoeva T, Boyanova L, Stankova P, Pencheva D, Keuleyan E, et al. Dissemination of successful international clone ST15 and clonal complex 17 among Bulgarian CTX-M-15 producing K. pneumoniae isolates. Diagnostic Microbiology and Infectious Disease. 2017 ;89(4):310-3. doi:10.1016/j.diagmicrobio.2017.08.012
- Khalifa AB, Khedher M. Epidémiologie des souches de Klebsiella spp. uropathogènes productrices de β-lactamases à spectre élargi dans un hôpital universitaire Tunisien, 2009. Pathologie Biologie. 2012 ;60(2):e1-5. doi:10.1016/j.patbio.2010.11.003
- Dameanti FN, Yanestria SM, Effendi MH, Plumeriastuti H, Tyasningsih W, Ugbo EN, et al. Identification of blaSHV and blaTEM extended spectrum beta-lactamase genes in Klebsiella pneumoniae in the dairy wastewater, East Java Province, Indonesia. Biodiversitas Journal of Biological Diversity. 2023;24(11): 6092-99. doi:10.13057/biodiv/d241130
- Espinar MJ, Rocha R, Ribeiro M, Gonçalves Rodrigues A, Pina-Vaz C. Extended-spectrum β-lactamases of Escherichia coli and Klebsiella pneumoniae screened by the VITEK 2 system. Journal of medical microbiology. 2011;60(6):756-60. doi:10.1099/jmm.0.024075-0
- Bora A, Hazarika NK, Shukla SK, Prasad KN, Sarma JB, Ahmed G. Prevalence of blaTEM, blaSHV and blaCTX-M genes in clinical isolates of Escherichia coli and Klebsiella pneumoniae from Northeast India. Indian Journal of Pathology and Microbiology. 2014 ;57(2):249-54. doi:10.4103/0377-4929.134698
- SALEEM M, RASHID F, FAIZ M. Genomic Analysis of Highly Virulent blaCTX-M, blaSHV and blaTEM Genes in Resistant Strains of E. coli and Klebsiella: an emerging threat. 2022; 16 (1). doi:10.53350/pjmhs22161186
- Eftekhar F, Rastegar M, Golalipoor M, Mansoursamaei N. Detection of Extended Spectrum B-Lactamases in Urinary Isolates of Klebsiella pneumoniae in Relation to blaSHV, blaTEM and blaCTX-M gene carriage. Iranian journal of public health. 2012;41(3):127-32. doi:N/A
- Pirzaman AN, Mojtahedi A. Investigation of antibiotic resistance and the presence of integron genes among ESBL producing Klebsiella isolates. Meta Gene. 2019; 19:37-41. doi:10.1016/j.mgene.2018.10.008
- Gravey F, Loggia G, de La Blanchardière A, Cattoir V. Bacterial epidemiology and antimicrobial resistance profiles of urinary specimens of the elderly. Medecine et maladies infectieuses. 2017 ;47(4):271-8. doi:10.1016/j.medmal.2017.03.002
- Marra AR, Wey SB, Castelo A, Gales AC, Cal RG, Filho JR, et al. Nosocomial bloodstream infections caused by Klebsiella pneumoniae: impact of extended-spectrum β-lactamase (ESBL) production on clinical outcome in a hospital with high ESBL prevalence. BMC infectious diseases. 2006; 6:1-8. doi:10.1186/1471-2334-6-24
- Ali FA, Ismael RM. Dissemination of Klebsiella pneumonia and Klebsiella oxytoca Harboring bla TEM genes isolated from different clinical samples in Erbil City. Diyala Journal of Medicine. 2017;12(2):40-51. doi:N/A
- Nirwati H, Sinanjung K, Fahrunissa F, Wijaya F, Napitupulu S, Hati VP, et al. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC proceedings. 2019; 13:1-8. doi:10.1186/s12919-019-0176-7
- Shahraki-Zahedani S, Rigi S, Bokaeian M, Ansari-Moghaddam A, Moghadampour M. First report of TEM-104-, SHV-99-, SHV-108-, and SHV-110-producing Klebsiella pneumoniae from Iran. Revista da Sociedade Brasileira de Medicina Tropical. 2016; 49:441-5. doi:10.1590/0037-8682-0114-2016
- Pishtiwan AH, Khadija KM. Prevalence of blaTEM, blaSHV, and blaCTX-M genes among ESBL-producing Klebsiella pneumoniae and Escherichia coli isolated from thalassemia patients in Erbil, Iraq. Mediterranean journal of hematology and infectious diseases. 2019;11(1):e2019041. doi:10.4084/mjhid.2019.041
- Ranjbar R, Fatahian Kelishadrokhi A, Chehelgerdi M. Molecular characterization, serotypes and phenotypic and genotypic evaluation of antibiotic resistance of the Klebsiella pneumoniae strains isolated from different types of hospital-acquired infections. Infection and drug resistance. 2019; 12:603-11. doi:N/A
- Muggeo A, Guillard T, Klein F, Reffuveille F, François C, Babosan A, et al. Spread of Klebsiella pneumoniae ST395 non-susceptible to carbapenems and resistant to fluoroquinolones in North-Eastern France. Journal of Global Antimicrobial Resistance. 2018; 13:98-103. doi:10.1016/j.jgar.2017.10.023
- Alibi S, Ferjani A, Boukadida J. Molecular characterization of extended spectrum beta-lactamases produced by Klebsiella pneumoniae clinical strains from a Tunisian Hospital. Medecine et maladies infectieuses. 2015;45(4):139-43. doi:10.1016/j.medmal.2015.01.010
- Paterson DL, Hujer KM, Hujer AM, Yeiser B, Bonomo MD, Rice LB, et al. Extended-spectrum β-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: dominance and widespread prevalence of SHV-and CTX-M-type β-lactamases. Antimicrobial agents and chemotherapy. 2003;47(11):3554-60. doi:10.1128/aac.47.11.3554-3560.2003
- Guessennd N, Bremont S, Gbonon V, Kacou-NDouba A, Ekaza E, Lambert T, et al. Résistance aux quinolones de type qnr chez les entérobactéries productrices de bêta-lactamases à spectre élargi à Abidjan en Côte d’Ivoire. Pathologie Biologie. 2008 ;56(7-8):439-46. doi:10.1016/j.patbio.2008.07.025

This work is licensed under a Creative Commons Attribution 4.0 International License.

